
CLS
Laboratory for Chemistry and Life Science, Institute of Integrated Research, Institute of Science Tokyo
東京科学大学
総合研究院
化学生命科学研究所

LAST UPDATE 2020/06/10
-
研究者氏名
Researcher Name朱博 Bo ZHU
助教 Assistant Professor -
所属
Affiliation東京科学大学 総合研究院 化学生命科学研究所
分子生命化学領域
Laboratory for Chemistry and Life Science, Institute of Integrated Research, Institute of Science Tokyo
Molecular bioscience -
研究キーワード
Research Keywords蛋白質工学
高速分子進化工学
無細胞蛋白質合成
Protein engineering
In vitro directed evolution
Cell-free protein synthesis
- 研究テーマ
Research Subject -
分子ディスプレイ法を用いた機能性タンパク質のエンジニアリングと評価
Engineering and profiling of functional proteins via biomolecular display technologies
研究の背景 Background of the Research
Functional proteins, including enzymes, antibodies, biosensors, regulatory factors, etc., play essential roles in living organisms. Many of these proteins have been or have the potential to be utilized in the chemical and food industry [1], diagnostics [2-4] (Fig. 1), medical treatment, and bioremediation. The biomolecular display technologies could serve as rapid and cost-efficient tools for better understanding, improving, and creating these functional proteins. The powerful in vitro display technologies I have been working on have their unique advantages in protein/peptide-related research. The phage display [3] or mRNA display (Fig. 2) can handle a million to trillion-level library and do a great job of finding the novel targets of interest and describing the fitness landscape of a selected function. The microbead display [5] (Fig. 3) performs better in evolving the existing functions in terms of substrate affinity and turnover.
研究の目標 Research Objective
A synergetic effect is expected when different display technologies and machine learning work in a consecutive or feedback manner. The primary focus of my current research is on developing versatile research platforms using phage/mRNA display, microbead display, and machine learning, and the applications on functional antibody conjugate engineering, enzyme engineering, novel enzyme discovery, and fitness landscape profiling.
研究図Figures
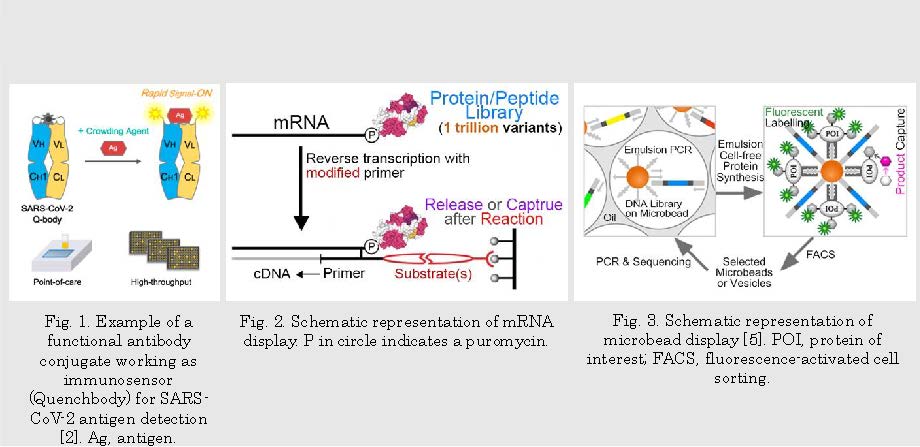
Fig. 2. Schematic representation of mRNA display. P in circle indicates a puromycin.
Fig. 3. Schematic representation of microbead display [5]. POI, protein of interest; FACS, fluorescence-activated cell sorting.
論文発表 / Publications
[1] Biosens. Bioelectron., 219, 114793 (2023); [2] Analyst, 147, 4971-4979 (2022); [3] Sci. Rep., 12, 15496 (2022); [4] Biochemistry, 62, 309-317 (2023); [5] PLoS ONE, 10, e0127479 (2015); [6] Analyst, 148, 1422-1429 (2023).
研究者連絡先 / HP
- zhu.b.ac
 m.titech.ac.jp、shu.h.424b
m.titech.ac.jp、shu.h.424b m.isct.ac.jp
m.isct.ac.jp - http://www.ueda.res.titech.ac.jp/
- https://kitaguchi.jimdofree.com/